Steven Ness@sness23544
is under improvement
49
Events attended
14
Submissions made
Canada
8+ years of experience
About me
Scientist and CEO
I built with
AudioCraftis under improvement
🤝 Top Collaborators
🤓 Latest Submissions

TechBio Lead Gen and SMYKM
**The $3.2B Problem We're Solving** Life sciences sales teams lose 80% of productivity to manual research. In TechBio alone, reps spend 3+ hours per prospect reading papers, analyzing research, and attempting personalization - yet achieve only 2% response rates. The culprit? Generic outreach that fails the "Show Me You Know Me" (SMYKM) test. **SMYKM AI: Intelligence at Scale** Our platform deploys four specialized AI agents via IBM watsonx Orchestrate: **Lead Discovery** scans publications, funding databases, and conference proceedings to identify prospects with urgent equipment needs. It scores leads based on research alignment, budget availability, and buying signals. **Profile Builder** analyzes each prospect's complete research footprint - papers, patents, presentations - extracting equipment requirements, technical bottlenecks, and validation challenges that create purchase urgency.**Product Match** maps prospect needs to your catalog, generating compelling comparisons that position your solution against alternatives. It identifies unique value propositions specific to their research goals. **SMYKM Outreach** synthesizes all intelligence into personalized messages that reference specific achievements, acknowledge real pain points, and position products as solutions to their exact challenges. **Product Match** maps prospect needs to your catalog, generating compelling comparisons that position your solution against alternatives. It identifies unique value propositions specific to their research goals. **SMYKM Outreach** synthesizes all intelligence into personalized messages that reference specific achievements, acknowledge real pain points, and position products as solutions to their exact challenges.
23 Nov 2025
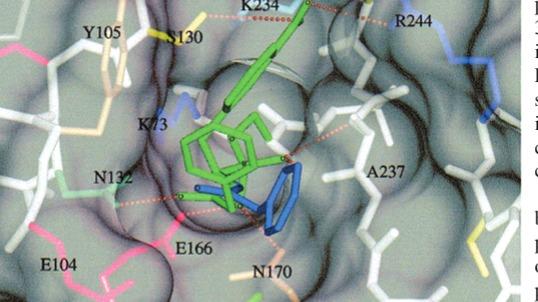
Plinderp - accellerating bio research
"Accelerating Research in Artificial Intelligence for Structural Biology." The project's main goal is to leverage Llama 3.2 to analyze and gain insights from scientific papers focused on molecular docking—specifically, how ligands bind to protein targets. Understanding the intricacies of protein-small molecule docking can reveal binding affinities and specific molecular interactions like hydrogen bonds and hydrophobic contacts. By analyzing these binding patterns, researchers can optimize ligand design for better therapeutic outcomes. PLINDER, a comprehensive dataset of 449,383 PLI systems, aims to enhance predictions in small molecule drug design by addressing limitations found in existing datasets, such as size and diversity. Despite its grandeur, there were gaps in citation information for certain structures, prompting the team to engage with the PLINDER team via Discord, who recommended accessing the Protein Data Bank (PDB). To address these gaps, the bioAI team presents "Plinderp," a website and database providing citations, metadata links to relevant papers, and sometimes full-text articles for all PLINDER entries. They also created a Python package, "plinderpdoibio," enhancing user access to the Plinder dataset. For the hackathon, the team developed a new website, plinderp.doi.bio, which aggregates citation links for PLINDER entries, allowing users to access valuable research information efficiently. A specific example is provided for the TEM beta-lactamase enzyme in Escherichia coli, showcasing its citation links and full-text availability. Additionally, an API is available for easy access to Plinderp functionalities. The Python package hosted on PyPi allows fast access to the original dataset for community use. Focusing on reducing barriers for Llama developers, the team highlights the complexities of working with large language models in structural biology, particularly in data preparation and evaluation.
11 Nov 2024

Asymptotic Cuteness
**Asymptotic Cuteness: The Infinite Cat Optimization Loop** is an innovative project that leverages advanced artificial intelligence models to iteratively enhance the cuteness of a cat video. Developed during the lablab.ai hackathon, the project utilizes rhymes.ai's state-of-the-art models, **Aria** and **Allegro**, to create a self-improving system based on reinforcement learning principles. The process begins with **Allegro**, an advanced generative model that creates videos from textual prompts. By inputting the simple prompt **"a cute cat,"** Allegro generates an initial video featuring an adorable feline. This video serves as the starting point for optimization. Next, **Aria**, a multimodal large language model capable of understanding and processing both text and images, analyzes the video. Aria evaluates various aspects such as the cat's facial features, expressiveness, fur softness, color vibrancy, and overall emotional impact. It provides a cuteness rating and suggests areas for enhancement. Key frames capturing the essence of cuteness are extracted from the video. Using Aria's capabilities, these frames are enhanced by focusing on elements that increase appeal—making the cat's eyes larger and more expressive, softening the fur texture, brightening colors, and adding playful poses or backgrounds. The enhanced images are then fed back into Allegro to generate a new, improved video. This forms an optimization loop where each iteration aims to produce a cuter video than the last, approaching the asymptote of ultimate cuteness. The process mirrors reinforcement learning: - **State**: The current version of the video. - **Action**: Enhancing images and regenerating the video. - **Reward**: The cuteness rating provided by Aria.
4 Nov 2024

BioX AI
there are approximately 100 million scientific papers ever published and of those 30 million papers are in biology. we propose to use artificial intelligence with retrieval augmented generation to solve various biological problems. With this vast amount of knowledge there are many different tasks that this could be applied to in biology. This is an enormous hidden gold mine, sitting in plain sight. there are many such problems in biology that we could attack, such as life extension, development of vaccines, treatment for rare diseases, development of new antibiotics and cancer. In the field of cancer, there are many different kinds of treatments, some more amenable to computational methods than others. Some of these treatments have the potential to be used for many different cancers, we call these platforms.
20 Oct 2024
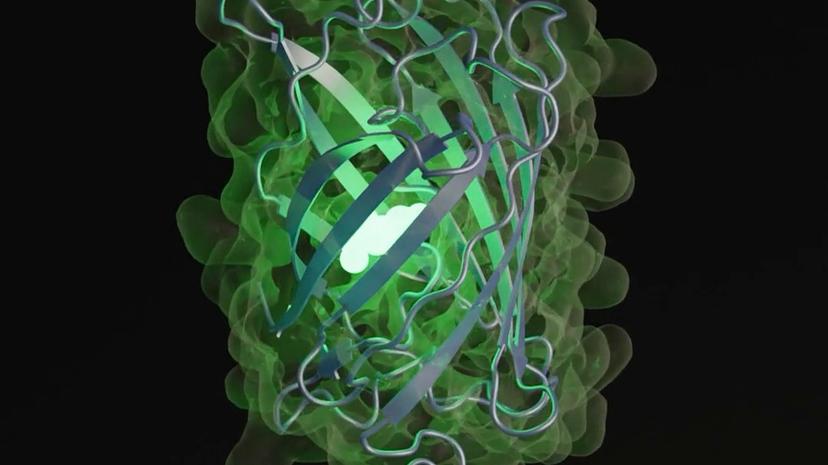
bioAI understanding Protein LLMs with AI
The bioAI team, led by Steven Ness, is exploring the innovative intersection of large language models (LLMs) and protein sequences, particularly focusing on the ESM3 (Evolutionary Scale Modelling) framework developed by a team from Meta AI. ESM3 is a cutting-edge Protein Large Language Model that applies LLM techniques to analyze and design green fluorescent proteins (GFPs) from the DNA sequence level. The team undertook a comprehensive examination of the ESM3 architecture, producing a detailed 68-page paper rich in technical content relevant to both artificial intelligence and structural biology. Utilizing scripts to process a markdown file generated from the original PDF, the team executed 24 diverse experiments that shed light on the efficacy and capabilities of ESM3. The results are openly shared on GitHub, alongside a series of markdown files and a YouTube video aimed at demystifying the findings for both expert and non-expert audiences. The project underscores the potential of AI in biotechnology, paving the way for advancements in protein engineering and molecular biology. By making these insights publicly available, the team aims to foster greater understanding and collaboration within the scientific community.
7 Aug 2024

AI to understand synthetic biology
Synthetic biology is a new field where we apply principles of engineering to biology. It has been responsible for some radical new advances in biotechnology. Some researchers and companies have been applying AI principles to biology with stunning effect. We want help accelerate progress in synthetic biology by using AI. In this project, we are looking at student work for the final group project in the joint MIT Media Lab and Harvard Class, How To Grow Almost Anything (https://htgaa.org). This final project involves making mutants of a virus specific to E.coli, the MS2 phage. We asked Claude3 Opus to first mark all the student reports. We also asked Claude3 to design it's own experiments, based on the work of the students.
16 Mar 2024

Accelerating synthetic biology with AI
Synthetic Biology is a new exciting field where we are applying engineering concepts to biology. Currently I am a Global TA for a class from MIT called "How To Grow Almost Anything" where 30 in person and over 100 global listeners learn from the leading researchers and companies in the field of synthetic biology, like George Church from Harvard and Emily Proust from Twist Biosciences. Our final project for this course involves phage therapy to treat multidrug resistant bacteria, like E. coli. We are focusing on the MS2 bacteriophage, a virus that only infects bacteria, and on the lysis protein in this bacteria. For this project, a detailed literature search would be desirable. A quick search on Entrez gives 1696 abstracts for the search query "ms2 and phage". We develop a python program to download and rate all the abstracts for their relevance to this project.
7 Mar 2024
AI Cancer Review Writer
Review articles in scientific journals are a helpful way for researchers to learn the state of the art in a field from leading names in the field. In the field of cancer treatment, there are many such review articles in the literature. One common issue with review articles is that they are almost immediately out of date, that is, as soon as a new article is published with new data that changes the scientific consensus about a problem, the review article will not have this information. In addition, many review articles are published in prestigious journals, which means that they are often expensive and thereby inaccessible to a large part of the world. We have sketched out an AI in this hackathon that writes up to date literature reviews from the very latest literature, downloaded fresh from Entrez PubMed, a vast archive of biological abstracts, with millions of peer-reviewed abstracts about cancer. We use this information along with Retrieval Augmented Generation to generate sentences for a new article, where each sentence is based on one from a chosen literature review article.
22 Jan 2024
biologyAI 3D movies
Your AI-driven tool is a revolutionary blend of Midjourney and Stable Video, designed specifically for science YouTubers. It simplifies the creation of educational videos by transforming static, microscopic world images into dynamic, 3D CGI-like videos, accompanied by AI-generated audio tracks. This innovation allows content creators to produce high-quality, engaging visuals effortlessly, focusing more on their educational narratives. The standout feature of your project is its capacity to automate the production of intricate, visually rich content. This not only makes the process efficient but also accessible to creators with varying levels of technical expertise. The focus on microscopic imagery offers a deep dive into the cellular world, providing viewers with an immersive and detailed exploration of scientific concepts. Another key aspect is the AI-generated audio tracks. These tracks complement the visuals, enhancing the overall viewer experience and engagement. This is particularly beneficial for YouTubers who seek to create a complete sensory experience for their audience. The potential of your tool extends far beyond its current application. It holds the promise of revolutionizing the production of educational content across various digital platforms. By making complex scientific topics visually appealing and easy to understand, your tool can play a crucial role in expanding the reach and impact of science communication. As part of an AI startup accelerator, your project stands out for its innovative use of AI, its impact on educational content quality, and its broad potential in enhancing the field of science education. This tool not only represents a significant advancement in content creation technology but also in the democratization of high-quality educational resources.
22 Dec 2023


.png&w=640&q=75)

















